Outputs
Gallery
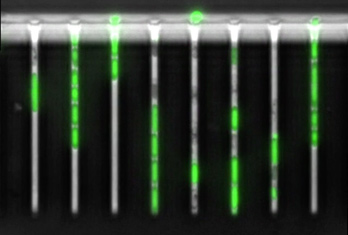
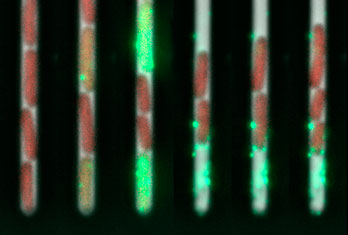
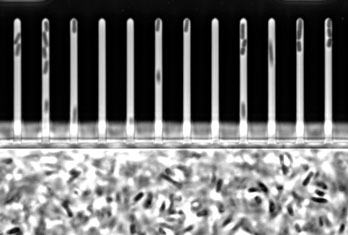
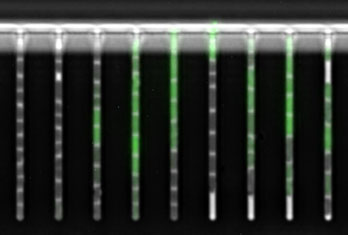
Let’s start with the fun bit: here are videos and pictures showcasing various aspects of my research projects.
Publications
This list might be neither exhaustive nor up-to-date: you can also find me on google scholar.
[preprint] Jacquel B, Kavčič B, Matifas A, Julou T & Charvin G (2023) A trade-off between stress resistance and tolerance underlies the adaptive response to hydrogen peroxide. Biorxiv 2021.04.23.440814.
doi:10.1101/2021.04.23.440814
doi:10.1101/2021.04.23.440814
[preprint] Julou T, Gervais T & van Nimwegen E (2022) Growth rate controls the sensitivity of gene regulatory circuits. Biorxiv 2022.04.03.486858.
doi:10.1101/2022.04.03.486858
doi:10.1101/2022.04.03.486858
Kolter R, Balaban N & Julou T (2022) Bacteria grow swiftly and live thriftily. Current Biology 32, R599–R605.
doi:10.1016/j.cub.2022.05.005
doi:10.1016/j.cub.2022.05.005
Sobota M et al. (2022) The expression of virulence genes increases membrane permeability and sensitivity to envelope stress in Salmonella Typhimurium. PLoS Biology 20, e3001608.
doi:10.1371/journal.pbio.3001608
doi:10.1371/journal.pbio.3001608
Urchueguía A, Galbusera L, Chauvin D, Bellement G, Julou T & van Nimwegen E (2021) Genome-wide gene expression noise in Escherichia coli is condition-dependent and determined by propagation of noise through the regulatory network. PLoS Biology 19, e3001491.
doi:10.1371/journal.pbio.3001491
doi:10.1371/journal.pbio.3001491
Julou T, Zweifel L, Blank D, Fiori A & van Nimwegen E (2020) Subpopulations of sensorless bacteria drive fitness in fluctuating environments. PLoS Biology 18, e3000952.
doi:10.1371/journal.pbio.3000952
Images (raw and preprocessed datasets), raw data and analysis scripts.
doi:10.1371/journal.pbio.3000952
Images (raw and preprocessed datasets), raw data and analysis scripts.
Galbusera L, Bellement-Theroue G, Urchueguia A, Julou T & van Nimwegen E (2020) Using fluorescence flow cytometry data for single-cell gene expression analysis in bacteria. PLoS One 15, e0240233.
doi:10.1371/journal.pone.0240233
doi:10.1371/journal.pone.0240233
Witz G, van Nimwegen E & Julou T (2019) Initiation of chromosome replication controls both division and replication cycles in E. coli through a double-adder mechanism. eLife 8, e48063. doi:10.7554/eLife.48063
Images and raw data with analysis scripts.
Our rebuttal to a comment of this work by the Jun's lab.
Images and raw data with analysis scripts.
Our rebuttal to a comment of this work by the Jun's lab.
Kaiser M, Jug F, Julou T, Deshpande S, Pfohl T, Silander OK, Myers G & Nimwegen E van (2018) Monitoring single-cell gene regulation under dynamically controllable conditions with integrated microfluidics and software. Nature Communications 9, 212.
doi:10.1038/s41467-017-02505-0
Images and raw data.
doi:10.1038/s41467-017-02505-0
Images and raw data.
Denoth Lippuner A, Julou T & Barral Y (2014) Budding yeast as a model organism to study the effects of age. FEMS Microbiol. Rev. 38, 300–325.
doi:10.1111/1574-6976.12060
doi:10.1111/1574-6976.12060
Julou T, Mora T, Guillon L, Croquette V, Schalk IJ, Bensimon D & Desprat N (2013) Cell-cell contacts confine public goods diffusion inside Pseudomonas aeruginosa clonal microcolonies. Proc. Natl. Acad. Sci. U.S.A. 110, 12577–12582.
doi:10.1073/pnas.1301428110
doi:10.1073/pnas.1301428110
Coquel A-S, Jacob J-P, Primet M, Demarez A, Dimiccoli M, Julou T, Moisan L,
Lindner AB & Berry H (2013) Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect. PLoS Comput. Biol. 9, e1003038.
doi:10.1371/journal.pcbi.1003038
doi:10.1371/journal.pcbi.1003038
Julou T, Desprat N, Bensimon D & Croquette V (2012) Monitoring microbial population dynamics at low densities. Rev Sci Instrum 83, 074301.
doi:10.1063/1.4729796
doi:10.1063/1.4729796
Flutre T, Julou T, Riboli-Sasco L & Ribrault C (2011) Research community: Pilot scheme for misconduct database. Nature 478, 37.
doi:10.1038/478037c
doi:10.1038/478037c
Julou T (2011) Evolution, competition and cooperation in bacterial populations. PhD thesis, Ecole Normale Supérieure (Paris).
link
link
Flutre T, Julou T, Riboli-Sasco L & Ribrault C (2010) Scientific Red Cards: a collaborative website for better communication between scientists and institutions about misconduct. European Science Editing 36, 49.
link
link
Mosconi F, Julou T, Desprat N, Sinha DK, Allemand J-F, Croquette V & Bensimon D (2008) Some nonlinear challenges in biology. Nonlinearity 21, T131.
doi:10.1088/0951-7715/21/8/T03
doi:10.1088/0951-7715/21/8/T03
Julou T, Burghardt B, Gebauer G, Berveiller D, Damesin C & Selosse M-A (2005) Mixotrophy in orchids: insights from a comparative study of green individuals and nonphotosynthetic individuals of Cephalanthera damasonium. The New phytologist 166, 639–653.
doi:10.1111/j.1469-8137.2005.01364.x
doi:10.1111/j.1469-8137.2005.01364.x
Talks & Posters
Qbio Seminar (2023) at ENS, Paris (France).
PoLS Seminar (2023) at EPFL, Lausanne (Switzerland).
Program “Quantitative Biology of Non-growing Microbes” (2022; Program Organizer) at KITP, Santa Barbara (USA).
4th International Workshop on Microbial Life under Exteme Energy Limitation (2022) at Sandbjerg Manor, Denmark.
Workshop “Major ideas in quantitative microbial physiology: Past, Present and Future” (2022) at Copenhagen (Denmark).
Cell Size and Growth Seminar (2022) online (hosted by Stanford).
Institute Seminar (2021) online (hosted by MPI Plön, Germany).
BIRS workshop “Advances in Theoretical and Experimental Methods for Analyzing Complex Regulatory Networks” (2020; Invited Talk) at BIRS (Banff, Canada). (video)
Séminaire du LiPhy (2019) at Univ. Grenoble Alpes (France).
q-bio Conference (2019) at San Francisco (USA).
Workshop “Information Processing in Single Cells” (2019) at Aspen Center for Physics (USA).
2nd SPP1617 International Conference "Phenotypic heterogeneity and sociobiology of bacterial populations" (2019; Invited Talk), at Schloss Hohenkammer (Germany).
EMBO workshop "Bacterial Persistence & Antimicrobial Therapy" (2018; Poster), at Ascona (Switzerland).
IS2M Annual Meetings: Microfluidics in Upper Rhine Valley (2018), at Université de Haute Alsace (France).
Institute Seminar (2018) at Institute of Cell Biology & SynthSys, University of Edinburgh (UK).
FriSBi seminar (2018) at IST (Austria).
Principles of microbial adaptation (2018; Poster) at Lorentz Center, Leiden (NL).
EMBO workshop “Optimizations and trade-offs in cellular growth and survival” (2018) at Weizmann Institute (Israel).
Quantitative Methods in Gene Regulation IV (2017) at Cambridge (UK).
Basel Computational Biology Conference [BC]2 (2017) at Basel (Switzerland).
Eco-Evolutionary Dynamics in Nature and the Lab (2017) at KITP, Santa Barbara (USA). (video)
New Approaches and Concepts in Microbiology (2017; Poster) at EMBL, Heidelberg (Germany).
Biozentrum Symposium (2017; Poster) at Basel (Switzerland).
Quantitative Laws Summer School (2016) at Como (Italy).
Microbiology Department Seminar (2016) at UNIGE, Genève (Switzerland).
Rigi Workshop (2016; Poster) at Luzern (Switzerland).
Biozentrum Symposium (2016; Poster) at Basel (Switzerland).
New Approaches and Concepts in Microbiology (2015; Poster) at EMBL, Heidelberg (Germany).
Experimental Approaches to Evolution & Ecology (2014; Poster) at EMBL, Heidelberg (Germany).
Computational Biology Seminar (2014) at ETH Zürich (Switzerland).
Institute Seminar (2013) at IGBMC, Strasbourg (France).
New Approaches and Concepts in Microbiology (2013; 2 Posters!!) at EMBL, Heidelberg (Germany).
Experimental Approaches to Evolution & Ecology (2012; Poster) at EMBL, Heidelberg (Germany).
Seminaire du LiPhy (2011) at Univ. Joseph Fourier, Grenoble (France).
Paris-Zürich Young Scientists Symposium in Applied Microscopy (2011) at ETH Zürich (Switzerland).
MMEMS conference “Evolution of Microbial Cooperation” (2011; Poster) at University of Bath (UK).
Between unicellularity and multicellularity: microbes in interaction (2010) at Institut des Systèmes Complexes, Paris (France).
The Evolution of Cooperation: Paradoxes of Collectivity & Individuality (2010) at Indiana University, Bloomington (USA).
102nd Statistical Mechanics Conference (2009) at Rutgers University (USA).
Journées du LPS (2008) at ENS, Paris (France).
New perspectives on the systematics & ecology of Orchids (2004) at Univ. P. Sabatier, Toulouse (France).
5ème Rencontres de Mycopathologie J. Chevaugeon (2004) at Aussois (France).